Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines
-
-
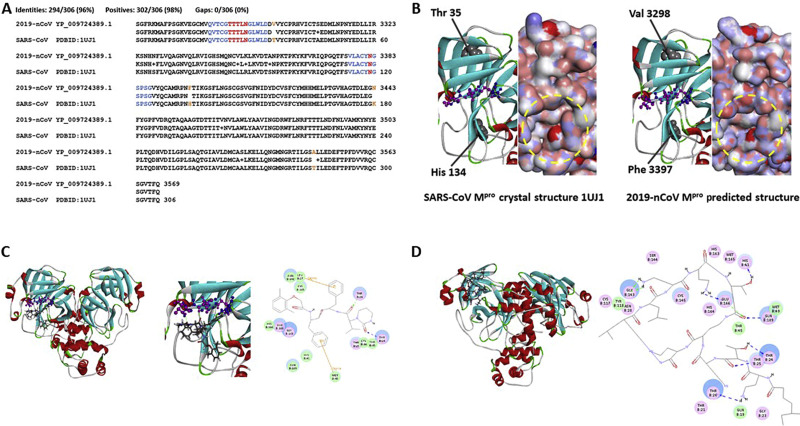
Fig. 1. Screen for potential 2019-nCoV Mpro inhibitors from commercial medicines. A: Sequence comparison between 2019-nCoV Mpro and SARS-Cov Mpro. Amino acids forming hydrogen bonds with drugs are shown in red, and their adjacent 5 amino acids on each side are shown in blue. Mutated amino acids rather than positive substitutions are shown in orange. B: Structural comparison of lopinavir/ritonavir binding pocket in SARS-CoV Mpro and 2019-nCoV Mpro. Ribbon models show the pocket structure of SARS-CoV Mpro (left) and 2019-nCoV Mpro (right), with α-helixes shown in red and β-sheets in cyan. Residues essential for lopinavir/ritonavir binding are shown in ball-and-stick format, of which Thr24, Thr26, and Asn28 are shown in purple, and Thr25, Leu27, and Asn119 are shown in blue. Protein solid surface model is shown to the right of each ribbon model, with the outer rim of the binding pocket marked by dashed yellow cycle. Mutations (marked in orange in panel A) are shown as gray balls, which are apart from the binding pocket. C: Docking model of lopinavir to 2019-nCoV Mpro. Left, overall docking model of lopinavir to 2019-nCoV Mpro; middle, enlargement of the lopinavir binding region; right, predicted chemical bonds between lopinavir and key residues of the binding pocket.D: Docking model of colistin to 2019-nCoV Mpro. Left, overall docking model of colistin to 2019-nCoV Mpro; right, predicted chemical bonds between colistin and key residues of the binding pocket. In (C) and (D), protein ribbon models are shown with the same diagram as described in (B), and drugs are shown as sticks. Hydrogen bonds between drugs and amino acids are shown as dash lines, and Pi bonds are shown as orange lines.
Table 1. Predicted commercial medicines as potential inhibitors against 2019-nCoV Mpro.
Name H-bond count Vital residues for H-bond formation Medical indications in DrugBank Colistin 9 THR24, THR25, THR26 Antibiotic Valrubicin 7 THR24, THR25, THR26,
ASN28, ASN119Anthracycline, antitumor Icatibant 6 ASN28, ASN119 Hereditary angioedema Bepotastine 5 THR25, THR26, ASN119 Rhinitis, uriticaria/puritus Epirubicin 4 ASN28, ASN119 Antitumor Epoprostenol 4 ASN119 Vasodilator, platelet aggregation Vapreotide 3 THR24, ASN28, ASN119 Antitumor Aprepitant 3 ASN28, ASN119 Nausea, vomiting, antitumor Caspofungin 3 ASN119 Antifungal Perphenazine 2 ASN28, ASN119 Antipsychotic -
[1] Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J.R., and Hilgenfeld, R., 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 300, 1763-1767. [2] Cui, J., Li, F., and Shi, Z.L., 2019. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 17, 181-192. [3] Dhama, K., Pawaiya, R., Chakraborty, S., Tiwari, R., M, S., and Verma, A., 2014. Coronavirus infection in equines: a review. Asian J. Anim. Vet. Adv. 9, 164-176. [4] Gorbalenya, A.E., Donchenko, A.P., Blinov, V.M., and Koonin, E.V., 1989. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold. FEBS Lett. 243, 103-114. [5] Hui, D.S., E, I.A., Madani, T.A., Ntoumi, F., Kock, R., Dar, O., Ippolito, G., McHugh, T.D., Memish, Z.A., Drosten, C., Zumla, A., and Petersen, E., 2020. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect Dis. 91, 264-266. [6] Lai, M.M.C., Holmes, K.V., 2001. Coronaviridae: the viruses and their replication, in: Knipe, D.M. and Howley, P.M. (Eds.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, pp. 1163-1179. [7] Lu, H., Stratton, C.W., and Tang, Y.W., 2020. Outbreak of pneumonia of unknown etiology in Wuhan China: the mystery and the miracle. J. Med. Virol. DOI: 10.1002/jmv.25678. [8] Masters, P.S., Perlman, S., 2013. Coronaviridae, in: Knipe, D.M. and Howley, P.M. (Eds.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, pp. 825-858. [9] Nukoolkarn, V., Lee, V.S., Malaisree, M., Aruksakulwong, O., and Hannongbua, S., 2008. Molecular dynamic simulations analysis of ritonavir and lopinavir as SARS-CoV 3CL(pro) inhibitors. J. Theor. Biol. 254, 861-867. [10] Regenmortel, M.H.V. van, Fauquet, C.M., Bishop, D.H.L., Carstens, E.B., Estes, M. K., Lemon, S.M., Maniloff, J., Mayo, M.A., McGeoch, D.J., Pringle, C.R., Wickner, R.B., 2000. Coronaviridae, in: Regenmortel, M.H.V. van, Fauquet, C.M., Bishop, D.H.L., et al. (Eds.), Virus Taxonomy: Classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, pp. 835-849. [11] Schoeman, D., and Fielding, B.C., 2019. Coronavirus envelope protein: current knowledge. Virol. J. 16, 69. [12] Xue, X., Yu, H., Yang, H., Xue, F., Wu, Z., Shen, W., Li, J., Zhou, Z., Ding, Y., Zhao, Q., Zhang, X.C., Liao, M., Bartlam, M., and Rao, Z., 2008. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 82, 2515-2527. [13] Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G.F., and Tan, W., 2020. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. DOI: 10.1056/NEJMoa2001017. -





 下载:
下载:

 京公网安备 11010502036328号 京ICP备17033152号
京公网安备 11010502036328号 京ICP备17033152号